CSSS 508, Week 3
Rebecca Ferrell
April 13, 2016
Weaning you off of spreadsheets
Today we'll talk about dplyr: a package that does in R just about any calculation you've tried to do in Excel, but more transparently, reproducibly, and safely. Don't be the sad research assistant who made this mistake (Reinhart and Rogoff):

Modifying data frames with dplyr
Filtering rows (subsetting)
Recall last week we used the filter command to subset data like so:
library(dplyr)
library(gapminder)
Canada <- gapminder %>%
filter(country == "Canada")
Excel analogue:

%in%
Common use case: want to filter rows to things in some set. The c() function (combine, concatenate) is how we make vectors in R, which is an important data type. (We'll talk more about those next week.)
former_yugoslavia <- c("Bosnia and Herzegovina", "Croatia", "Macedonia", "Montenegro", "Serbia", "Slovenia")
yugoslavia <- gapminder %>%
filter(country %in% former_yugoslavia)
tail(yugoslavia, 2)
Source: local data frame [2 x 6]
country continent year lifeExp pop gdpPercap
(fctr) (fctr) (int) (dbl) (int) (dbl)
1 Slovenia Europe 2002 76.660 2011497 20660.02
2 Slovenia Europe 2007 77.926 2009245 25768.26
What values are out there? Use distinct
You can see all the values in your data for columns using distinct:
gapminder %>% distinct(continent, year)
Source: local data frame [60 x 6]
country continent year lifeExp pop gdpPercap
(fctr) (fctr) (int) (dbl) (int) (dbl)
1 Afghanistan Asia 1952 28.801 8425333 779.4453
2 Afghanistan Asia 1957 30.332 9240934 820.8530
3 Afghanistan Asia 1962 31.997 10267083 853.1007
4 Afghanistan Asia 1967 34.020 11537966 836.1971
5 Afghanistan Asia 1972 36.088 13079460 739.9811
6 Afghanistan Asia 1977 38.438 14880372 786.1134
7 Afghanistan Asia 1982 39.854 12881816 978.0114
8 Afghanistan Asia 1987 40.822 13867957 852.3959
9 Afghanistan Asia 1992 41.674 16317921 649.3414
10 Afghanistan Asia 1997 41.763 22227415 635.3414
.. ... ... ... ... ... ...
Time to talk about pipes (%>%)
The dplyr package makes use of an operator (not native to R) called a pipe. We write pipes like %>% (Cntl-Shift-M). Pipes take the object on the left and apply the function on the right: x %>% f(y) = f(x, y). Read out loud: “and then…”
gapminder %>% filter(country == "Canada") %>% head(2)
Source: local data frame [2 x 6]
country continent year lifeExp pop gdpPercap
(fctr) (fctr) (int) (dbl) (int) (dbl)
1 Canada Americas 1952 68.75 14785584 11367.16
2 Canada Americas 1957 69.96 17010154 12489.95
Pipes save us typing and allow chaining like above, so we use them all the time when manipulating data frames.
Piping
- Pipes are clearer to read when you have each function on a separate line (inconsistent in these slides because of space constraints).
take_this_data %>%
do_first_thing(with = this_value) %>%
do_next_thing(using = that_value) %>% ...
Stuff to the left of the pipe is passed to the first argument of the function on the right. Other arguments go on the right in the function.
If you ever find yourself piping a function where the data is not the first argument, use
.in the data argument instead.
yugoslavia %>% lm(pop ~ year, data = .)
Sampling rows: sample_n
We can also filter at random to work with a smaller dataset using sample_n or sample_frac.
set.seed(0413) # makes random numbers repeatable
yugoslavia %>% sample_n(size = 6, replace = FALSE)
Source: local data frame [6 x 6]
country continent year lifeExp pop gdpPercap
(fctr) (fctr) (int) (dbl) (int) (dbl)
1 Montenegro Europe 1962 63.728 474528 4649.594
2 Montenegro Europe 1982 74.101 562548 11222.588
3 Serbia Europe 1962 64.531 7616060 6289.629
4 Slovenia Europe 1952 65.570 1489518 4215.042
5 Serbia Europe 1952 57.996 6860147 3581.459
6 Croatia Europe 1997 73.680 4444595 9875.605
Sorting: arrange
Along with filtering the data to see certain rows, we might want to sort it:
yugoslavia %>% arrange(year, desc(pop))
Source: local data frame [60 x 6]
country continent year lifeExp pop gdpPercap
(fctr) (fctr) (int) (dbl) (int) (dbl)
1 Serbia Europe 1952 57.996 6860147 3581.4594
2 Croatia Europe 1952 61.210 3882229 3119.2365
3 Bosnia and Herzegovina Europe 1952 53.820 2791000 973.5332
4 Slovenia Europe 1952 65.570 1489518 4215.0417
5 Montenegro Europe 1952 59.164 413834 2647.5856
6 Serbia Europe 1957 61.685 7271135 4981.0909
7 Croatia Europe 1957 64.770 3991242 4338.2316
8 Bosnia and Herzegovina Europe 1957 58.450 3076000 1353.9892
9 Slovenia Europe 1957 67.850 1533070 5862.2766
10 Montenegro Europe 1957 61.448 442829 3682.2599
.. ... ... ... ... ... ...
Keeping columns: select
Not only can we limit rows, but we can limit columns (and put them in the order listed) using select.
yugoslavia %>% select(country, year, pop) %>% head(4)
Source: local data frame [4 x 3]
country year pop
(fctr) (int) (int)
1 Bosnia and Herzegovina 1952 2791000
2 Bosnia and Herzegovina 1957 3076000
3 Bosnia and Herzegovina 1962 3349000
4 Bosnia and Herzegovina 1967 3585000
Dropping columns: select
We can instead drop columns with select using - signs:
yugoslavia %>% select(-continent, -pop, -lifeExp) %>% head(4)
Source: local data frame [4 x 3]
country year gdpPercap
(fctr) (int) (dbl)
1 Bosnia and Herzegovina 1952 973.5332
2 Bosnia and Herzegovina 1957 1353.9892
3 Bosnia and Herzegovina 1962 1709.6837
4 Bosnia and Herzegovina 1967 2172.3524
Helper functions for select
select() has a variety of helper functions like starts_with, ends_with, and contains, or giving a range of continguous columns startvar:endvar. These are very useful if you have a “wide” data frame with column names following a pattern or ordering. See ?select.
(US Dept. of Education “College Scorecard” data: > 100 columns)
Renaming columns with select
We can rename columns using select, but that drops everything that isn't mentioned:
yugoslavia %>%
select(Life_Expectancy = lifeExp) %>%
head(4)
Source: local data frame [4 x 1]
Life_Expectancy
(dbl)
1 53.82
2 58.45
3 61.93
4 64.79
Safer: rename columns with...rename
yugoslavia %>%
select(country, year, lifeExp) %>%
rename(Life_Expectancy = lifeExp) %>%
head(4)
Source: local data frame [4 x 3]
country year Life_Expectancy
(fctr) (int) (dbl)
1 Bosnia and Herzegovina 1952 53.82
2 Bosnia and Herzegovina 1957 58.45
3 Bosnia and Herzegovina 1962 61.93
4 Bosnia and Herzegovina 1967 64.79
Column naming practices
Good column names will be self-describing. Don't use inscrutable abbreviations to save typing. RStudio's autocompleting functions take away the pain of long variable names: hit tab while writing code.
Valid “naked” column names can contain upper or lowercase letters, numbers, periods, and underscores. They must start with a letter or period and not be a special reserved word (e.g.
TRUE,if).Names are case-sensitive:
Yearandyearare not the same thing!You can include spaces or use reserved words if you put backticks around the name. Spaces can be worth including when preparing data for
ggplot2orpandersince you don't have to rename axes or table headings.
Column name with space example
library(pander)
yugoslavia %>% filter(country == "Serbia") %>%
select(year, lifeExp) %>%
rename(Year = year, `Life Expectancy` = lifeExp) %>%
head(5) %>%
pander(style = "rmarkdown", caption = "Serbian life expectancy")
| Year | Life Expectancy |
|---|---|
| 1952 | 57.996 |
| 1957 | 61.685 |
| 1962 | 64.531 |
| 1967 | 66.914 |
| 1972 | 68.700 |
Table: Serbian life expectancy
Create new columns: mutate and transmute
Thing you do in spreadsheets: add column to data, drag down.

dplyr way: add new columns to a data frame using mutate. (Add new columns and drop old ones using transmute.)
mutate example
yugoslavia %>% filter(country == "Serbia") %>%
select(year, pop, lifeExp) %>%
mutate(pop_million = pop / 1000000,
life_exp_past_40 = lifeExp - 40) %>%
head(5)
Source: local data frame [5 x 5]
year pop lifeExp pop_million life_exp_past_40
(int) (int) (dbl) (dbl) (dbl)
1 1952 6860147 57.996 6.860147 17.996
2 1957 7271135 61.685 7.271135 21.685
3 1962 7616060 64.531 7.616060 24.531
4 1967 7971222 66.914 7.971222 26.914
5 1972 8313288 68.700 8.313288 28.700
ifelse()
A common function used in mutate (and in general in R programming) is ifelse(). This returns a value depending on logical tests.
yugoslavia %>%
mutate(short_country = ifelse(country == "Bosnia and Herzegovina", "B and H", as.character(country))) %>%
select(short_country, year, pop) %>%
arrange(year, short_country) %>%
head(3)
Source: local data frame [3 x 3]
short_country year pop
(chr) (int) (int)
1 B and H 1952 2791000
2 Croatia 1952 3882229
3 Montenegro 1952 413834
Summarizing with dplyr
General aggregation: summarize
summarize takes your rows of data and computes something across them: count how many rows there are, calculate the mean or total, etc. You can use any function that aggregates multiple values into a single one (like sd).
In a spreadsheet:
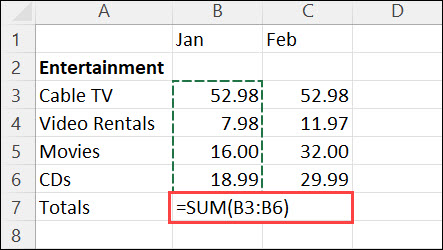
Summarize example
yugoslavia %>%
filter(year == 1982) %>%
summarize(n_obs = n(),
total_pop = sum(pop),
mean_life_exp = mean(lifeExp),
range_life_exp = max(lifeExp) - min(lifeExp))
Source: local data frame [1 x 4]
n_obs total_pop mean_life_exp range_life_exp
(int) (int) (dbl) (dbl)
1 5 20042685 71.2952 3.939
Avoiding repetition: summarize_each
Maybe you need to calculate the mean and standard deviation of a bunch of columns. With summarize_each(), you put the functions to use in a funs() list, and the variables to compute over after that (like select syntax).
yugoslavia %>%
filter(year == 1982) %>%
summarize_each(funs(mean, sd),
lifeExp, pop)
Source: local data frame [1 x 4]
lifeExp_mean pop_mean lifeExp_sd pop_sd
(dbl) (dbl) (dbl) (dbl)
1 71.2952 4008537 1.602685 3237282
Splitting data into groups: group_by
The special function group_by() changes how functions operate on the data, most importantly summarize. These functions are computed within each group as defined by variables given, rather than over all rows at once. Typically the variables you group by will be integers, factors, or characters, and not continuous real values.
Excel analogue: pivot tables
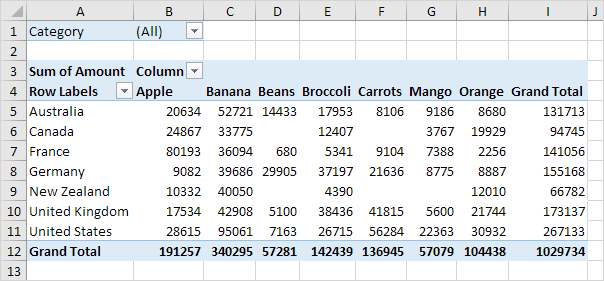
group_by() example
yugoslavia %>% group_by(year) %>%
summarize(num_countries = n_distinct(country),
total_pop = sum(pop),
total_gdp_per_cap = sum(pop * gdpPercap) / total_pop) %>%
head(5)
Source: local data frame [5 x 4]
year num_countries total_pop total_gdp_per_cap
(int) (int) (int) (dbl)
1 1952 5 15436728 3029.794
2 1957 5 16314276 4187.491
3 1962 5 17099107 5256.578
4 1967 5 17878535 6655.827
5 1972 5 18579786 8730.215
Window functions
Grouping can also be used with mutate or filter to give rank orders within a group, lagged values, and cumulative sums. Much more on window functions is in a vignette.
yugoslavia %>% select(country, year, pop) %>%
filter(year >= 2002) %>% group_by(country) %>%
mutate(lag_pop = lag(pop, order_by = year),
pop_chg = pop - lag_pop) %>% head(4)
Source: local data frame [4 x 5]
Groups: country [2]
country year pop lag_pop pop_chg
(fctr) (int) (int) (int) (int)
1 Bosnia and Herzegovina 2002 4165416 NA NA
2 Bosnia and Herzegovina 2007 4552198 4165416 386782
3 Croatia 2002 4481020 NA NA
4 Croatia 2007 4493312 4481020 12292
Lab break!
With the Gapminder data, practice the following analyses:
- Find the population (in units of millions of people) in 2007 for countries in Asia. Make a histogram.
- Use population and GDP per capita to find the 10 largest economies in 1952 as measured by overall GDP (not per capita).
- For each country and each year, find the percentage growth of the population relative to the population of the country 5 years prior (the Gapminder data are reported every 5 years). Then find the 10 country-year pairs that had the highest percentage growth relative to 5 years prior, and the 10 country-year pairs that had the lowest percentage growth.
Joining data frames
When do we need to join tables?
- Want to make columns using criteria too complicated for
ifelse() - Combine data stored in separate places: e.g. UW registrar information with student homework grades
Excel equivalents: VLOOKUP, MATCH

Joining: conceptually
We need to think about the following when we want to merge data frames A and B:
- Which rows are we keeping from each data frame?
- Which columns are we keeping from each data frame?
- Which variables determine whether rows match?
Types of joins: rows and columns to keep
A %>% inner_join(B): keep rows fromAthat match rows inB, columns from bothAandBA %>% left_join(B): keep all rows fromA, matched withBwherever possible (NAwhen not), columns from bothAandBA %>% right_join(B): keep all rows fromB, matched withAwherever possible (NAwhen not), columns from bothAandBA %>% full_join(B): keep all rows from eitherAorB, matched wherever possible (NAwhen not), columns from bothAandBA %>% semi_join(B): keep rows fromAthat match rows inB, columns from justAA %>% anti_join(B): keep rows fromAthat don't match a row inB, columns from justA
Matching criteria
We say rows should match because they have some columns containing the same value. We list these in a by = argument to the join.
- No
by: matches using all variables inAandBthat have identical names by = c("var1", "var2", "var3"): matches on identical values ofvar1,,var2,var3in bothAandBby = c("Avar1" = "Bvar1", "Avar2" = "Bvar2"): matches identical values ofAvar1variable inAtoBvar1variable inB, andAvar2variable inAtoBvar2variable inB
Note: if there are multiple matches, you'll get one row for each possible combo (except with semi_join and anti_join).
(Need to get more complicated? You'll want to learn SQL.)
nycflights13 data
We'll use data in the nycflights13 package. Install and load it:
# install.packages("nycflights13")
library(nycflights13)
It includes five tables, some of which contain missing data (NA):
flights: flights leaving JFK, LGA, or EWR in 2013airlines: airline abbreviationsairports: airport metadataplanes: airplane metadataweather: hourly weather data for JFK, LGA, and EWR
Join example #1
Who manufactures the planes that flew to Seattle?
flights %>% filter(dest == "SEA") %>% select(tailnum) %>%
left_join(planes %>% select(tailnum, manufacturer), by = "tailnum") %>%
distinct(manufacturer)
Source: local data frame [6 x 2]
tailnum manufacturer
(chr) (chr)
1 N594AS BOEING
2 N503JB AIRBUS INDUSTRIE
3 N3ETAA NA
4 N712JB AIRBUS
5 N508JB CIRRUS DESIGN CORP
6 N531JB BARKER JACK L
Join example #2
Which airlines had the most flights to Seattle from NYC?
flights %>% filter(dest == "SEA") %>% select(carrier) %>%
left_join(airlines, by = "carrier") %>%
group_by(name) %>% tally() %>% arrange(desc(n))
Source: local data frame [5 x 2]
name n
(fctr) (int)
1 Delta Air Lines Inc. 1213
2 United Air Lines Inc. 1117
3 Alaska Airlines Inc. 714
4 JetBlue Airways 514
5 American Airlines Inc. 365
Join example #3
Is there a relationship between departure delays and wind gusts?
library(ggplot2)
flights %>% select(origin, year, month, day, hour, dep_delay) %>%
inner_join(weather, by = c("origin", "year", "month", "day", "hour")) %>%
select(dep_delay, wind_gust) %>%
# removing rows with missing values
filter(!is.na(dep_delay) & !is.na(wind_gust)) %>%
ggplot(aes(x = wind_gust, y = dep_delay)) +
geom_point() + geom_smooth()
Wind gusts and delays
Redo after removing extreme outliers, just trend
flights %>% select(origin, year, month, day, hour, dep_delay) %>%
inner_join(weather, by = c("origin", "year", "month", "day", "hour")) %>%
select(dep_delay, wind_gust) %>%
filter(!is.na(dep_delay) & !is.na(wind_gust) & wind_gust < 250) %>%
ggplot(aes(x = wind_gust, y = dep_delay)) +
geom_smooth() + theme_bw(base_size = 16) +
xlab("Wind gusts in departure hour (mph)") +
ylab("Average departure delay (minutes)")
Wind gusts and delays: mean trend
Lab break!
Some possible questions to investigate:
- What are the names of the most common destination airports?
- Which airlines fly from NYC to your home city?
- Is there a relationship between departure delays and precipitation?
- Use the time zone data in
airportsto convert flight arrival times to NYC local time.- What is the distribution of arrival times for flights leaving NYC over a 24 hour period?
- Are especially late or early arrivals particular to some regions or airlines?
Warning! flights has 336776 rows, so if you do a sloppy join, you can end up with many matches per observation and have the data blow up.
Homework
Pick something to look at in the nycflights13 data and write up a .Rmd file showing your investigation. Upload both the .Rmd file and the .html file to Canvas. You must use at least once: mutate, summarize, group_by, and joins. Include at least one formatted plot or table.
This time, include all your code in your output document, using comments and line breaks separating commands so that it is clear to a peer what you are doing. You must write up your observations in words as well.
Grading rubric
- 0: Didn't turn anything in
- 1: Turned in but low effort, ignoring many directions
- 2: Decent effort, followed directions with some minor issues
- 3: Nailed it